SHP099 is a small molecule inhibitor of SHP2 discovered by Novartis. ReadCrystal has replicated the structure of the
small molecule-protein complex of SHP099 with SHP2 to a resolution of 1.9 Å.
Background
SHP2 (Src homology 2 containing protein tyrosine phosphatase2) is a non-receptor protein tyrosine phosphatase
(PTP) encoded by the PTPN11 gene. SHP2 plays an important role as a phosphatase in maintaining the homeostasis of
protein tyrosine phosphorylation and is involved in the regulation of several signaling pathways (RAS/RAF/MEK/ERK,
PI3K/AKT and JAK/STAT). Due to its involvement in the regulation of several important signaling pathways, the
over-activation of SHP2 is associated with a variety of cancers. Therefore, it has become an important target for
cancer drug development.
SHP2 contains two SH2 structural domains (N-SH2, C-SH2), a highly conserved
PTP catalytic domain and a C-terminal tail region containing 2 tyrosine phosphorylation sites (Tyr542 and Tyr580).
In the non-activated state, N-SH2 of SHP2 binds to the PTP structural domain to maintain the autoinhibited state
of phosphatase activity; when SH2 binds to the phosphorylated tyrosine residues, the protein conformation is
changed and the autoinhibited state is lifted to perform the function of phosphatase to dephosphorylate the target
protein.
As a potential target for cancer therapy, the development of SHP2 inhibitors has become one of
the popular areas for antitumor drug development. The traditional idea of SHP2 inhibitor design is to target its
orthosteric site (PTP catalytic active center), however, due to PTP is high conserved, the small molecule
inhibitors designed for the catalytic structural domain of PTP have low selectivity. Another idea is to develop
allosteric inhibitors, and SHP099, developed by Novartis in 2016, is the first allosteric inhibitors for the SHP2
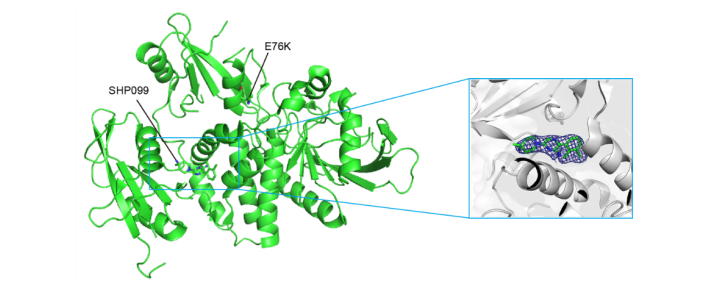
target. SHP099 binds to a tunnel-like allosteric pocket at the junction of the C-SH2, N-SH2 and PTP structural
domains, which maintains SHP2 in an autoinhibited conformation and inhibits catalytic activity.
Results
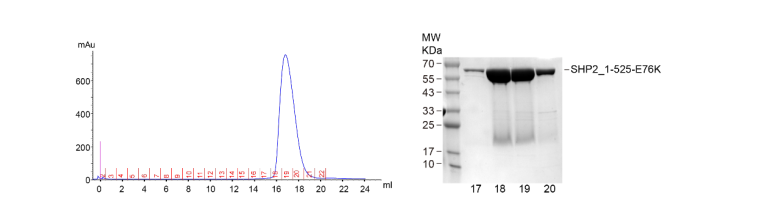
1. Protein expression & purification
The expression system used in this study was the E. coli expression system (BL21), and then Ni-NTA column was
used for protein purification followed by gel filtration chromatography.
Fig. 1 Results of gel filtration chromatography and SDS-PAGE
2. Protein crystallization and diffraction data collection
In this experiment, the protein crystals were crystallized by the sitting-drop method, and diffraction data
information was collected by synchrotron radiation source.
Fig. 2 SHP099-SHP2 eutectic and diffraction image
3. Structure determination
Determination of the three-dimensional structure of the SHP099-SHP2 complex with a resolution of 1.9 Å.
Fig. 3 Three-dimensional structure of SHP099-SHP2 complex