IRED is an enzyme important in industrial catalysis. The IRED in this case was obtained by semi-rational design based on structural biology data. Since there was no structure with high homology for reference, ReadCrystal delivered the structure data of this enzyme by de novo phasing.
Background
Imine reductases (IREDs) are NAPDH-dependent enzymes that catalyze the asymmetric reduction of imines to synthesize chiral amines. However, IREDs have a narrow range of substrates and so far rational engineering is rare.
In 2022, Shushan Gao's team at the Institute of Microbiology, Chinese Academy of Sciences published a paper on the structural design of imine reductase IR-G36 in the leading international academic journal 《Angewandte Chemie》. Through a combination of rational cavity design, combined active site saturation test (CAST) and thermostability engineering, the weakly active IR-G36 was transformed into the variant M5 with excellent performance and a 4193-fold improvement in catalytic efficiency. Moreover, M5 exhibits broad substrate scope for the synthesis of diverse azacycloalkylamines, and the reaction was demonstrated on a hectogram-scale under industrially relevant conditions.
The article performed enzyme modification by a semi-rational design approach. Firstly, the IR-G36 with the best R-selectivity (78%) was screened among 86 IREDs as a target for the next evolutionary step. Then structural biology was applied to determine the spatial structure of IR-G36 as the basis for enzyme modification, and finally the variant M5 with much higher catalytic efficiency was successfully modified.
The work on the structure determination of IREDs in this study was technically supported by ReadCrystal.
IREDs, imine reductases, belong to proteins, and the protein structure determination generally has the following steps:
-
Results
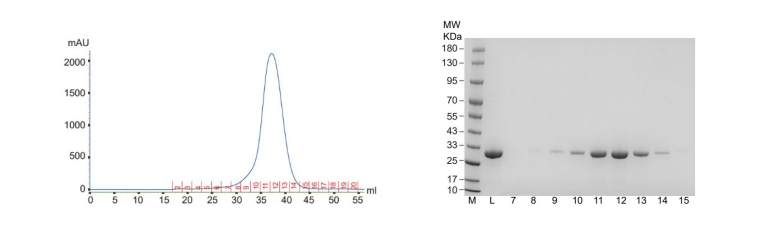
1. Protein expression & purification
The expression system used for IR-G36 protein expression in this study was the E.
coli
expression system (BL21), and then Ni-NTA column was used for protein purification followed by
gel filtration
chromatography.
Fig. 1 Results of gel filtration chromatography and SDS-PAGE
-
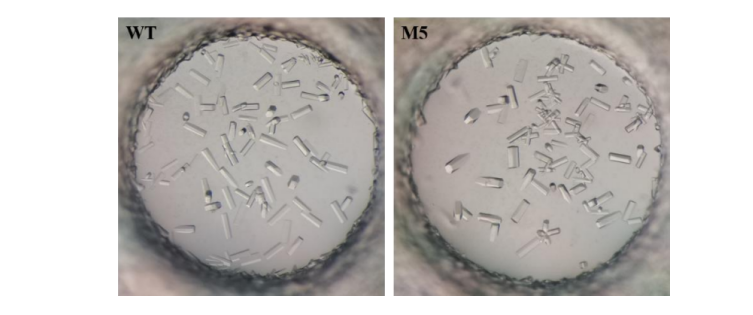
2. Protein crystallization
IR-G36 WT and IR-G36 M5 were crystallized by the sitting-drop method.
Fig. 2 Crystals of WT and M5 of IR-G36
-
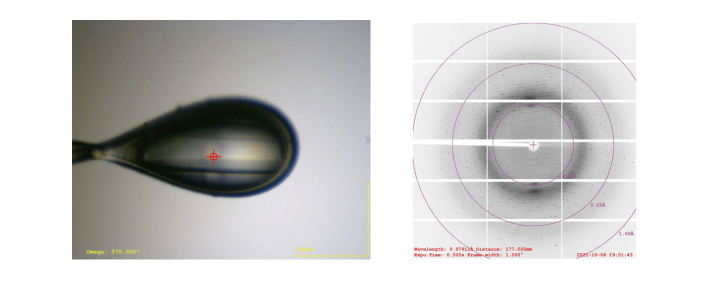
3. Diffraction data collection by X-ray
Diffraction data were collected by synchrotron radiation source.
Fig. 3 Diffraction image
-
4. Structure determination
Since IR-G36 is 40% homologous to the resolved IREDs, it is necessary to
using the method for de novo phasing. Heavy atom (Se atom) crystal derivatives were prepared and
the structure of IR-G36 WT was determined by single wavelength anomalous dispersion (SAD). The
resolution was 2.4 Å.
Fig. 4 Three-dimensional structure of WT IR-G36 in complex with cofactor NADP+
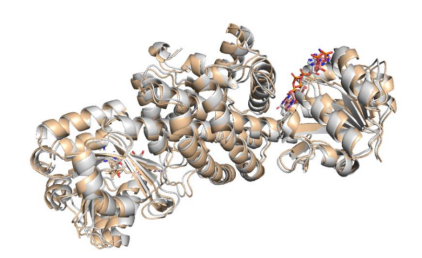
The structure of IR-G36 WT was used as a model to determine the structure of
IR-G36 M5 using the molecular replacement method.
Fig. 5 Overlay of the structures of IR-G36-WT (gray) with that of IR-G36-M5
(wheat)
-
Publication