-
+86 18962587269
-
contact@readcrystal.com
-
Changshu High-tech Industrial Development Zone, Suzhou, Jiangsu Province

On July 4th, Junliang Sun, the founder of ReadCrystal Technology, collaborated with a team of professors from Peking University to publish their latest research results in "Angew.Chem.Int.Ed." (IF 16.6). Their pioneering work utilized Cryo-3D electron diffraction, commonly referred to as MicroED, to determine the position of hydrogen atoms in COF materials. This significant achievement marks yet another remarkable breakthrough in the application of MicroED technology for advancing crystal structure analysis.

The synthesis of large single crystals for COF materials presents significant challenges, limiting the application of single-crystal X-ray diffraction in elucidating their crystal structure. The localization of hydrogen atoms is even more complex. Existing methods for locating hydrogen atoms, such as anisotropic refinement, powder X-ray diffraction, and single-crystal neutron diffraction, all demand higher standards in crystal and data quality. Three-dimensional electron diffraction (3D ED) has unique advantages for determining COF structure due to the robust electron-matter Coulomb interaction. This interaction enables the acquisition of high signal-to-noise electron diffraction data from nanocrystals. Using kinetic refinement techniques, it becomes possible to directly locate the position of all ordered atoms, including hydrogen atoms with guest molecules. This study pioneers the use of 3D ED to confirm the position of hydrogen atoms on guest molecules in COF materials.
Summary
Covalent organic frameworks (COFs) have extensive applications, their efficacy is deeply intertwined with host-guest interactions. Precisely locating all atoms, especially hydrogen, in COFs is pivotal for understanding these interactions. However, the complex process of synthesizing high-quality single crystals poses a challenge in determining hydrogen atom positions. Three-dimensional electron diffraction (3D ED) presents a breakthrough in resolving nanocrystals structures and identifying lighter atoms. This study marks a pioneering achievement by showing that continuous rotation electron diffraction tomography (cPEDT) through 3D ED, conducted under low-temperature conditions, enables the localization of hydrogen atoms not just within the framework but also on guest molecules within COFs. This novel approach sheds light on host-guest interactions by precisely mapping the positions of hydrogen atoms. These revelations offer a fresh perspective, significantly enhancing our understanding of COFs and their functionalities.
Abstract
Content preview
COF-300 samples with vacuum pores are denoted as COF-300-V, while COF-300 samples prepared in water, methanol, and ethanol are denoted as COF-300-H2O, COF-300-CH3OH, and COF-300-C2H5OH, respectively.
Position of hydrogen atoms in COF-300-V:
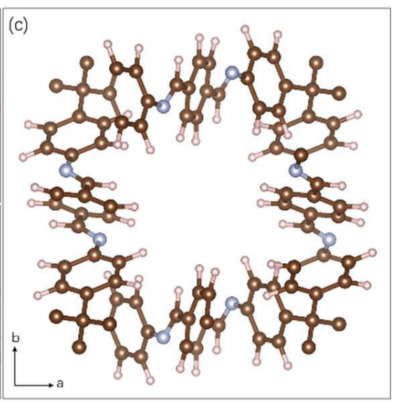
The 3D ED data of COF-300-V has a resolution higher than 0.85 Å, with a Rint of approximately 13.64%. The data quality is very close to that of SCXRD. The final structure of one channel of COF-300-V was obtained through dynamic refinement (hydrogen, white; carbon, brown; nitrogen, blue).
Position of hydrogen atoms in COF-300-H2O:

The cPEDT data resolution of the COF-300-H2O sample is higher than 0.8 Å, with an Rint value of 16.29%. The representation of the COF-300-H2O structure obtained through dynamic refinement along [001] is shown (hydrogen, white; carbon, brown; nitrogen, blue; oxygen, red; dashed lines represent hydrogen bonding interactions).

Interactions between host and guest molecules in the COF300-H2O channel. Water molecules form an ordered chain in the pores.
Positions of hydrogen atoms and disordered carbon atoms in COF-300-CH3OH and COF-300-C2H5OH:

COF300-CH3OH has a 3D ED data resolution higher than 0.85 Å, while COF-300-C2H5OH has a 3D ED data resolution higher than 0.9 Å. The final structural models of COF-300-CH3OH and COF-300-C2H5OH were obtained through dynamic refinement (hydrogen, white; carbon, brown; nitrogen, blue; oxygen, red. Dashed lines represent hydrogen bonds, and colored spheres represent partially occupied sites).
Various solvents adsorbed on COF-300 and their host-guest interactions.

Based on the obtained structural model, the main interactions between the host and guest molecules are hydrogen bonds between the nitrogen atom and hydroxyl group (Nframework···H—O), hydrogen bonds between the framework hydrogen atoms and water oxygen atoms (Hframework···OH2), and van der Waals interactions. These hydrogen bonds have moderate strength. The interactions between water molecules are stronger than those between methanol and ethanol molecules. These different interactions explain the phenomena observed during the sample preparation process.
Conclusion
For the first time, 3D ED was used to locate hydrogen atoms in the COF skeleton and guest molecules. Ordered hydrogen atoms can be easily located through kinetic refinement. In addition, kinematic refinement of cPEDT data revealed most of the hydrogen atom positions, which was due to the further reduction of dynamic effects and high data quality. Locating hydrogen atoms elucidated the host-guest interactions in COF-300. Considering the complexity of COF single crystal growth and the importance of host-guest interactions, we believe that the cPEDT method plays a key role in this field. The results of this study may help people understand various aspects of COFs and provide powerful tools for studying framework materials with water collection, catalysis, and proton transfer functions.
+86 18962587269
contact@readcrystal.com
Changshu High-tech Industrial Development Zone, Suzhou, Jiangsu Province